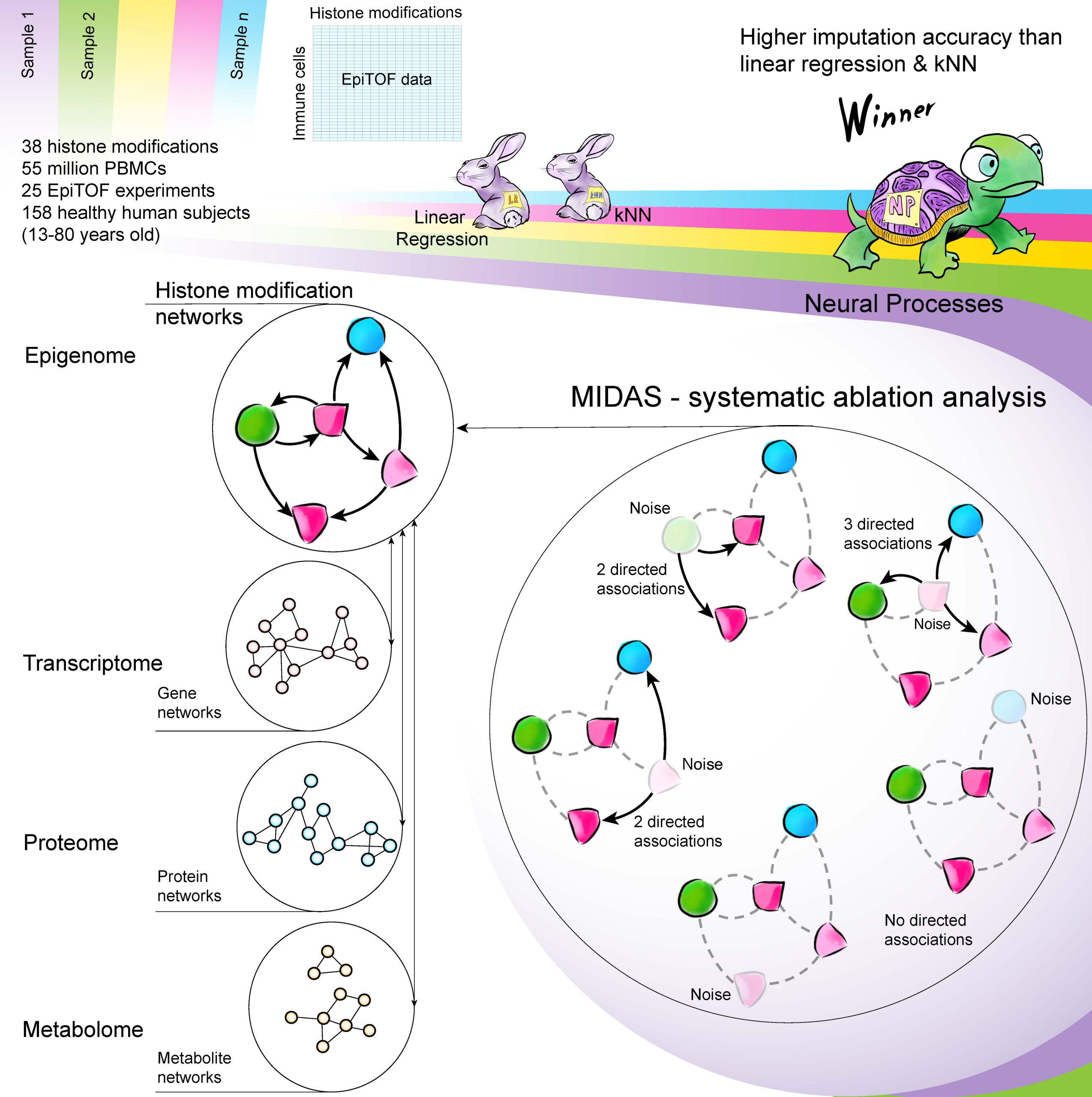
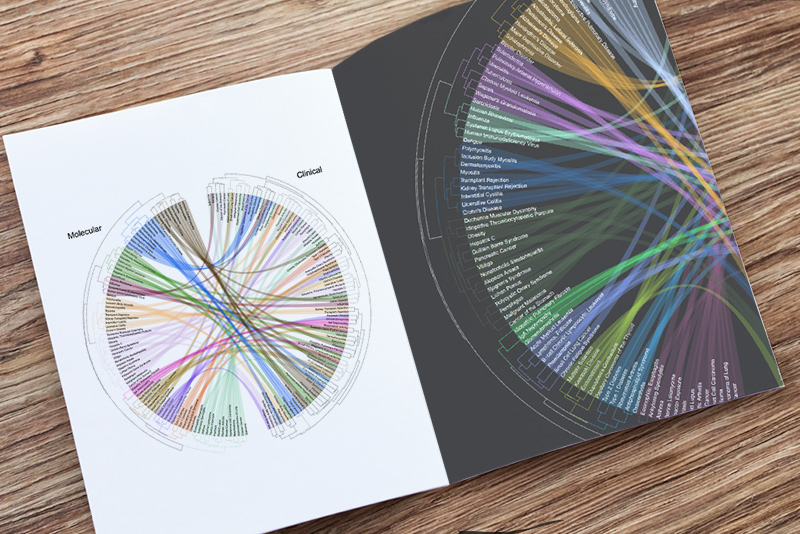
A typical biological experiment is a controlled experiment that explicitly limits biological, clinical, and technical heterogeneity. Although these controlled experiments have improved our understanding of biology, a controlled experiment does not represent the real-world heterogeneity. Therefore, its results are nearly impossible to translate into clinical practice immediately.
The Khatri Lab has developed a multi-cohort analysis framework for integrating these “dirty data” in a consistent manner that is representative of the biological, clinical, and technical heterogeneity observed in the real-world patient population. We have shown that compared to single-cohort analysis, following general guidelines of our framework, implemented in R packages MetaIntegrator or bayesMetaIntegrator, significantly improves reproducibility by integrating data from multiple independent cohorts, even when controlling for sample size. We have repeatedly demonstrated that utility of our framework in a broad spectrum of diseases including organ transplant rejection, infectious diseases (sepsis, bacterial infections, viral infections, tuberculosis, dengue), autoimmune diseases (systemic sclerosis, IBD, lupus), cancers (lung cancer, KRAS-associated cancers), pan-organ fibrosis, and vaccination for identifying signatures that are diagnostic, prognostic, therapeutic, and mechanistic.